The ineqx package implements Rosche (202X). [...]
Usage
ineqx(
treat = NULL,
post = NULL,
y,
ystat = "Var",
group = NULL,
time = NULL,
weights = NULL,
controls = NULL,
decomp = "post",
ref = NULL,
AME_mu = NULL,
AME_sigma = NULL,
dat
)Arguments
- treat
Character string. Treatment variable. Values must be 0/1
- post
Character string. Before/after variable. Values must be 0/1
- y
Character string. Dependent variable. Variable must be continuous.
- ystat
Character string. Either "Var" (default) or "CV2". Choose to analyze (the effect of x on) the variance or squared coefficient of variation.
- group
Character string. Variable must be categorical. Grouping variable to decompose variance into within- and between-group components.
- time
Character string. c.x with specify penalized splines, i.x will specify dummies. Time variable to analyze change over time.
- weights
Character string. Weight variable.
- controls
Character vector with additional control variables. E.g. c("c.age", "i.sex", ...)
- decomp
Character string. Either "post" (default) or "effect".
- ref
Number, vector, or list. Counterfactual reference point. See details.
- AME_mu
Dataframe with average marginal effects (Mu)
- AME_sigma
Dataframe with average marginal effects (Sigma)
- dat
Dataframe
Details
The main function of ineqx. [...]
Counterfactual reference point (ref)
ref can either be a number, c(number, "variable name"), or list(n=c(...), mu=c(...), sigma=c(...), beta=c(...), lambda=c(...)). [...]
Return
...
Examples
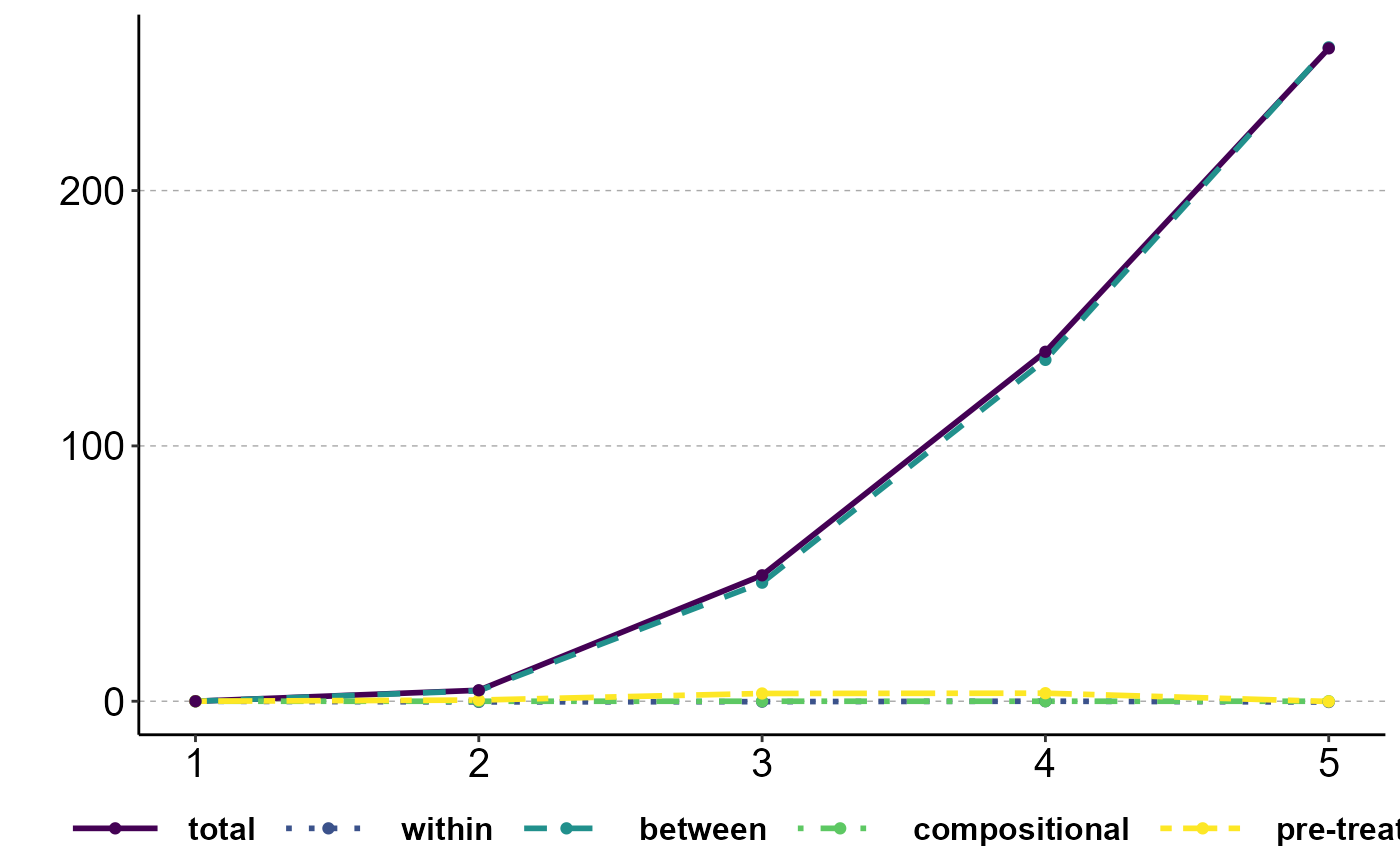
data(incdat)
# Descriptive variance decomposition
decomp_descr <- ineqx(y="inc", group="group", time="i.year", ref=1, dat=incdat)
#> Performing decomposition ...
#> Done.
plot(decomp_descr, type="dT")
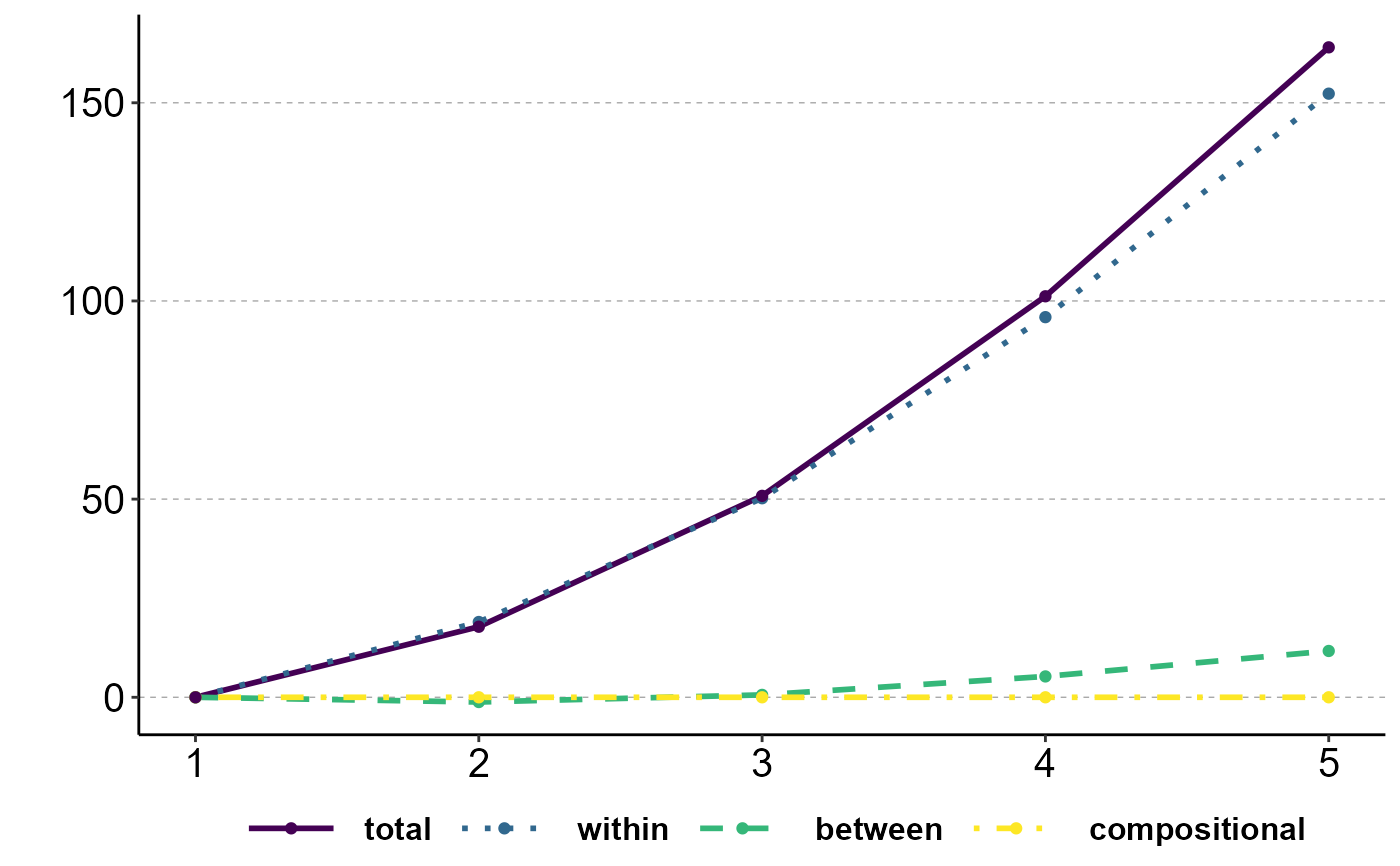 # Causal variance decomposition
decomp_treat <- ineqx(treat="x", post="t", y="inc", group="group", time="i.year", ref=1, dat=incdat)
#> Running variance function regression ...
#> GAMLSS-RS iteration 1: Global Deviance = 8406.392
#> GAMLSS-RS iteration 2: Global Deviance = 8406.348
#> GAMLSS-RS iteration 3: Global Deviance = 8406.348
#> Computing average marginal effects ...
#> Performing decomposition ...
#> Done.
plot(decomp_treat, type="dT")
# Causal variance decomposition
decomp_treat <- ineqx(treat="x", post="t", y="inc", group="group", time="i.year", ref=1, dat=incdat)
#> Running variance function regression ...
#> GAMLSS-RS iteration 1: Global Deviance = 8406.392
#> GAMLSS-RS iteration 2: Global Deviance = 8406.348
#> GAMLSS-RS iteration 3: Global Deviance = 8406.348
#> Computing average marginal effects ...
#> Performing decomposition ...
#> Done.
plot(decomp_treat, type="dT")